Upgma
UPGMA (unweighted pair group method with arithmetic mean) is a simple agglomerative (bottom-up) hierarchical clustering method.
It also has a weighted variant, WPGMA, and they are generally attributed to Sokal and Michener.
Note that the unweighted term indicates that all distances contribute equally to each average that is computed and does not refer to the math by which it is achieved. Thus the simple averaging in WPGMA produces a weighted result and the proportional averaging in UPGMA produces an unweighted result (see the working example).
Algorithm
The UPGMA algorithm constructs a rooted tree (dendrogram) that reflects the structure present in a pairwise similarity matrix (or a dissimilarity matrix). At each step, the nearest two clusters are combined into a higher-level cluster. The distance between any two clusters 








In other words, at each clustering step, the updated distance between the joined clusters 




The UPGMA algorithm produces rooted dendrograms and requires a constant-rate assumption - that is, it assumes an ultrametric tree in which the distances from the root to every branch tip are equal. When the tips are molecular data (i.e., DNA, RNA and protein) sampled at the same time, the ultrametricity assumption becomes equivalent to assuming a molecular clock.
Working example
This working example is based on a JC69 genetic distance matrix computed from the 5S ribosomal RNA sequence alignment of five bacteria: Bacillus subtilis (




First step
- First clustering
Let us assume that we have five elements 

| a | b | c | d | e | |
|---|---|---|---|---|---|
| a | 0 | 17 | 21 | 31 | 23 |
| b | 17 | 0 | 30 | 34 | 21 |
| c | 21 | 30 | 0 | 28 | 39 |
| d | 31 | 34 | 28 | 0 | 43 |
| e | 23 | 21 | 39 | 43 | 0 |
In this example, 



- First branch length estimation
Let 










- First distance matrix update
We then proceed to update the initial distance matrix 








Italicized values in 
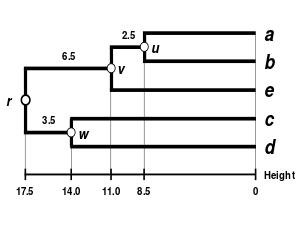
Second step
- Second clustering
We now reiterate the three previous steps, starting from the new distance matrix 
| (a,b) | c | d | e | |
|---|---|---|---|---|
| (a,b) | 0 | 25.5 | 32.5 | 22 |
| c | 25.5 | 0 | 28 | 39 |
| d | 32.5 | 28 | 0 | 43 |
| e | 22 | 39 | 43 | 0 |
Here, 



- Second branch length estimation
Let 








We deduce the missing branch length: 
- Second distance matrix update
We then proceed to update 





Thanks to this proportional average, the calculation of this new distance accounts for the larger size of the 


Proportional averaging therefore gives equal weight to the initial distances of matrix 
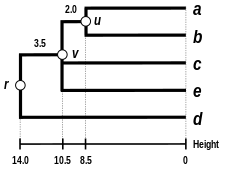
Third step
- Third clustering
We again reiterate the three previous steps, starting from the updated distance matrix 
| ((a,b),e) | c | d | |
|---|---|---|---|
| ((a,b),e) | 0 | 30 | 36 |
| c | 30 | 0 | 28 |
| d | 36 | 28 | 0 |
Here, 



- Third branch length estimation
Let 






- Third distance matrix update
There is a single entry to update, keeping in mind that the two elements 



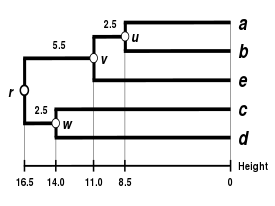
Final step
The final 
| ((a,b),e) | (c,d) | |
|---|---|---|
| ((a,b),e) | 0 | 33 |
| (c,d) | 33 | 0 |
So we join clusters 

Let 






We deduce the two remaining branch lengths:


The UPGMA dendrogram
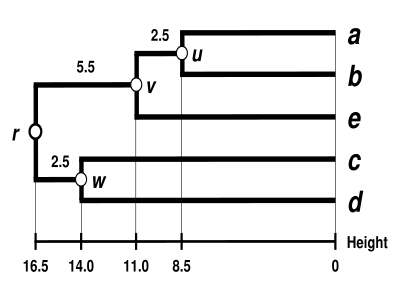
The dendrogram is now complete. It is ultrametric because all tips (



The dendrogram is therefore rooted by 
Comparison with other linkages
Alternative linkage schemes include single linkage clustering, complete linkage clustering, and WPGMA average linkage clustering. Implementing a different linkage is simply a matter of using a different formula to calculate inter-cluster distances during the distance matrix update steps of the above algorithm. Complete linkage clustering avoids a drawback of the alternative single linkage clustering method - the so-called chaining phenomenon, where clusters formed via single linkage clustering may be forced together due to single elements being close to each other, even though many of the elements in each cluster may be very distant to each other. Complete linkage tends to find compact clusters of approximately equal diameters.
 |  |  |  |
| Single-linkage clustering | Complete-linkage clustering | Average linkage clustering: WPGMA | Average linkage clustering: UPGMA. |
Uses
- In ecology, it is one of the most popular methods for the classification of sampling units (such as vegetation plots) on the basis of their pairwise similarities in relevant descriptor variables (such as species composition). For example, it has been used to understand the trophic interaction between marine bacteria and protists.
- In bioinformatics, UPGMA is used for the creation of phenetic trees (phenograms). UPGMA was initially designed for use in protein electrophoresis studies, but is currently most often used to produce guide trees for more sophisticated algorithms. This algorithm is for example used in sequence alignment procedures, as it proposes one order in which the sequences will be aligned. Indeed, the guide tree aims at grouping the most similar sequences, regardless of their evolutionary rate or phylogenetic affinities, and that is exactly the goal of UPGMA
- In phylogenetics, UPGMA assumes a constant rate of evolution (molecular clock hypothesis) and that all sequences were sampled at the same time, and is not a well-regarded method for inferring relationships unless this assumption has been tested and justified for the data set being used. Notice that even under a 'strict clock', sequences sampled at different times should not lead to an ultrametric tree.
Time complexity
A trivial implementation of the algorithm to construct the UPGMA tree has 


See also
References
External links
This article uses material from the Wikipedia English article UPGMA, which is released under the Creative Commons Attribution-ShareAlike 3.0 license ("CC BY-SA 3.0"); additional terms may apply (view authors). Content is available under CC BY-SA 4.0 unless otherwise noted. Images, videos and audio are available under their respective licenses.
®Wikipedia is a registered trademark of the Wiki Foundation, Inc. Wiki English (DUHOCTRUNGQUOC.VN) is an independent company and has no affiliation with Wiki Foundation.
