Filoviridae
Filoviridae (/ˌfaɪloʊˈvɪrɪdiː/) is a family of single-stranded negative-sense RNA viruses in the order Mononegavirales.
Two members of the family that are commonly known are Ebola virus and Marburg virus. Both viruses, and some of their lesser known relatives, cause severe disease in humans and nonhuman primates in the form of viral hemorrhagic fevers.
| Filoviridae | |
|---|---|
 | |
| Ebolavirus structure and genome | |
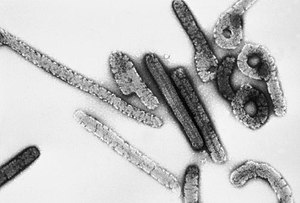 | |
| Electron micrograph of Marburg virus | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Monjiviricetes |
| Order: | Mononegavirales |
| Family: | Filoviridae |
| Genera | |
| |
All filoviruses are classified by the US as select agents, by the World Health Organization as Risk Group 4 Pathogens (requiring Biosafety Level 4-equivalent containment), by the National Institutes of Health/National Institute of Allergy and Infectious Diseases as Category A Priority Pathogens, and by the Centers for Disease Control and Prevention as Category A Bioterrorism Agents, and are listed as Biological Agents for Export Control by the Australia Group.
Use of term
The family Filoviridae is a virological taxon that was defined in 1982 and emended in 1991, 1998, 2000, 2005, 2010 and 2011. The family currently includes the six virus genera Cuevavirus, Dianlovirus, Ebolavirus, Marburgvirus, Striavirus, and Thamnovirus and is included in the order Mononegavirales. The members of the family (i.e. the actual physical entities) are called filoviruses or filovirids. The name Filoviridae is derived from the Latin noun filum (alluding to the filamentous morphology of filovirions) and the taxonomic suffix -viridae (which denotes a virus family).
Note
According to the rules for taxon naming established by the International Committee on Taxonomy of Viruses (ICTV), the name Filoviridae is always to be capitalized, italicized, never abbreviated, and to be preceded by the word "family". The names of its members (filoviruses or filovirids) are to be written in lower case, are not italicized, and used without articles.
Life cycle


The filovirus life cycle begins with virion attachment to specific cell-surface receptors, followed by fusion of the virion envelope with cellular membranes and the concomitant release of the virus nucleocapsid into the cytosol. The viral RNA-dependent RNA polymerase (RdRp, or RNA replicase) partially uncoats the nucleocapsid and transcribes the genes into positive-stranded mRNAs, which are then translated into structural and nonstructural proteins. Filovirus RdRps bind to a single promoter located at the 3' end of the genome. Transcription either terminates after a gene or continues to the next gene downstream. This means that genes close to the 3' end of the genome are transcribed in the greatest abundance, whereas those toward the 5' end are least likely to be transcribed. The gene order is therefore a simple but effective form of transcriptional regulation. The most abundant protein produced is the nucleoprotein, whose concentration in the cell determines when the RdRp switches from gene transcription to genome replication. Replication results in full-length, positive-stranded antigenomes that are in turn transcribed into negative-stranded virus progeny genome copies. Newly synthesized structural proteins and genomes self-assemble and accumulate near the inside of the cell membrane. Virions bud off from the cell, gaining their envelopes from the cellular membrane they bud from. The mature progeny particles then infect other cells to repeat the cycle.
Family inclusion criteria

A virus that fulfills the criteria for being a member of the order Mononegavirales is a member of the family Filoviridae if:
- it causes viral hemorrhagic fever in certain primates
- it infects primates, pigs or bats in nature
- it needs to be adapted through serial passage to cause disease in rodents
- it exclusively replicates in the cytoplasm of a host cell
- it has a genome ≈19 kbp in length
- it has an RNA genome that constitutes ≈1.1% of the virion mass
- its genome has a molecular weight of ≈4.2×106
- its genome contains one or more gene overlaps
- its genome contains seven genes in the order 3'-UTR-NP-VP35-VP40-GP-VP30-VP24-L-5'-UTR
- its VP24 gene is not homologous to genes of other mononegaviruses
- its genome contains transcription initiation and termination signals not found in genomes of other mononegaviruses
- it forms nucleocapsids with a buoyant density in CsCl of ≈1.32 g/cm3
- it forms nucleocapsids with a central axial channel (≈10–15 nm in width) surrounded by a dark layer (≈20 nm in width) and an outer helical layer (≈50 nm in width) with a cross striation (periodicity of ≈5 nm)
- it expresses a class I fusion glycoprotein that is highly N- and O-glycosylated and acylated at its cytoplasmic tail
- it expresses a primary matrix protein that is not glycosylated
- it forms virions that bud from the plasma membrane
- it forms virions that are predominantly filamentous (U- and 6-shaped) and that are ≈80 nm in width, and several hundred nm and up to 14 μm in length
- it forms virions that have surface projections ≈7 nm in length spaced ≈10 nm apart from each other
- it forms virions with a molecular mass of ≈3.82×108; an S20W of at least 1.40; and a buoyant density in potassium tartrate of ≈1.14 g/cm3
- it forms virions that are poorly neutralized in vivo
Family organization
| Genus name | Species name | Virus name (abbreviation) |
|---|---|---|
| Cuevavirus | Lloviu cuevavirus | Lloviu virus (LLOV) |
| Dianlovirus | Mengla dianlovirus | Měnglà virus (MLAV) |
| Ebolavirus | Bombali ebolavirus | Bombali virus (BOMV) |
| Bundibugyo ebolavirus | Bundibugyo virus (BDBV; previously BEBOV) | |
| Reston ebolavirus | Reston virus (RESTV; previously REBOV) | |
| Sudan ebolavirus | Sudan virus (SUDV; previously SEBOV) | |
| Taï Forest ebolavirus | Taï Forest virus (TAFV; previously CIEBOV) | |
| Zaire ebolavirus | Ebola virus (EBOV; previously ZEBOV) | |
| Marburgvirus | Marburg marburgvirus | Marburg virus (MARV) |
| Ravn virus (RAVV) | ||
| Striavirus | Xilang striavirus | Xīlǎng virus (XILV) |
| Thamnovirus | Huangjiao thamnovirus | Huángjiāo virus (HUJV) |
Phylogenetics
The mutation rates in these genomes have been estimated to be between 0.46 × 10−4 and 8.21 × 10−4 nucleotide substitutions/site/year. The most recent common ancestor of sequenced filovirus variants was estimated to be 1971 (1960–1976) for Ebola virus, 1970 (1948–1987) for Reston virus, and 1969 (1956–1976) for Sudan virus, with the most recent common ancestor among the four species included in the analysis (Ebola virus, Tai Forest virus, Sudan virus, and Reston virus) estimated at 1000–2100 years. The most recent common ancestor of the Marburg and Sudan species appears to have evolved 700 and 850 years before present respectively. Although mutational clocks placed the divergence time of extant filoviruses at ~10,000 years before the present, dating of orthologous endogenous elements (paleoviruses) in the genomes of hamsters and voles indicated that the extant genera of filovirids had a common ancestor at least as old as the Miocene (~16–23 million or so years ago).
Filoviridae cladogram is the following:
| Filoviridae |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Paleovirology
Filoviruses have a history that dates back several tens of million of years. Endogenous viral elements (EVEs) that appear to be derived from filovirus-like viruses have been identified in the genomes of bats, rodents, shrews, tenrecs, tarsiers, and marsupials. Although most filovirus-like EVEs appear to be pseudogenes, evolutionary analyses suggest that orthologs isolated from several species of the bat genus Myotis have been maintained by selection.
Vaccines
There are presently very limited vaccines for known filovirus. An effective vaccine against EBOV, developed in Canada, was approved for use in 2019 in the US and Europe. Similarly, efforts to develop a vaccine against Marburg virus are under way.
Mutation concerns and pandemic potential
There has been a pressing concern that a very slight genetic mutation to a filovirus such as EBOV could result in a change in transmission system from direct body fluid transmission to airborne transmission, as was seen in Reston virus (another member of genus Ebolavirus) between infected macaques. A similar change in the current circulating strains of EBOV could greatly increase the infection and disease rates caused by EBOV. However, there is no record of any Ebola strain ever having made this transition in humans.
The Department of Homeland Security’s National Biodefense Analysis and Countermeasures Center considers the risk of a mutated Ebola virus strain with aerosol transmission capability emerging in the future as a serious threat to national security and has collaborated with the Centers for Disease Control and Prevention (CDC) to design methods to detect EBOV aerosols.
References
External links


- ICTV Report: Filoviridae
- "Filoviridae". NCBI Taxonomy Browser. 11266.
- "FILOVIR". Scientific resources for research on filoviruses. Archived from the original on 2020-07-30. Retrieved 2014-08-08.
- Theoretical Evidence That The Ebola Virus Zaire Strain May Be Selenium-Dependent: A Factor In Pathogenesis And Viral Outbreaks? Taylor 1995
- Can Selenite Be An Ultimate Inhibitor Of Ebola And Other Viral Infections? Lipinski 2015 Archived 2020-11-19 at the Wayback Machine
- Many In West Africa May Be Immune To Ebola Virus New York Times
This article uses material from the Wikipedia English article Filoviridae, which is released under the Creative Commons Attribution-ShareAlike 3.0 license ("CC BY-SA 3.0"); additional terms may apply (view authors). Content is available under CC BY-SA 4.0 unless otherwise noted. Images, videos and audio are available under their respective licenses.
®Wikipedia is a registered trademark of the Wiki Foundation, Inc. Wiki English (DUHOCTRUNGQUOC.VN) is an independent company and has no affiliation with Wiki Foundation.