Genetics Deletion
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication.
Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur, which result in the deletion of a part of the chromosome. The breaks can be induced by heat, viruses, radiation, or chemical reactions. When a chromosome breaks, if a part of it is deleted or lost, the missing piece of chromosome is referred to as a deletion or a deficiency.
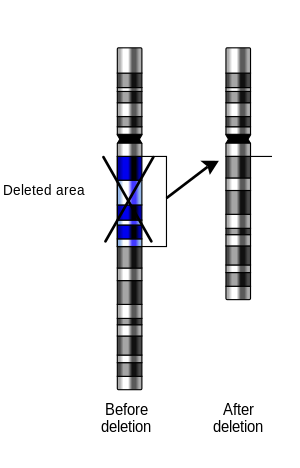
For synapsis to occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into a deletion or compensation loop.
The smallest single base deletion mutations occur by a single base flipping in the template DNA, followed by template DNA strand slippage, within the DNA polymerase active site.
Deletions can be caused by errors in chromosomal crossover during meiosis, which causes several serious genetic diseases. Deletions that do not occur in multiples of three bases can cause a frameshift by changing the 3-nucleotide protein reading frame of the genetic sequence. Deletions are representative of eukaryotic organisms, including humans and not in prokaryotic organisms, such as bacteria.
Causes
Causes include the following:
- Losses from translocation
- Chromosomal crossovers within a chromosomal inversion
- Unequal crossing over
- Breaking without rejoining
Types
Types of deletion include the following:
- Terminal deletion – a deletion that occurs towards the end of a chromosome.
- Intercalary/interstitial deletion – a deletion that occurs from the interior of a chromosome.
- Microdeletion – a relatively small amount of deletion (up to 5Mb that could include a dozen genes).
Micro-deletion is usually found in children with physical abnormalities. A large amount of deletion would result in immediate abortion (miscarriage).
Nomenclature

The International System for Human Cytogenomic Nomenclature (ISCN) is an international standard for human chromosome nomenclature, which includes band names, symbols and abbreviated terms used in the description of human chromosome and chromosome abnormalities. Abbreviations include a minus sign (−) for chromosome deletions, and del for deletions of parts of a chromosome.
Effects
Small deletions are less likely to be fatal; large deletions are usually fatal – there are always variations based on which genes are lost. Some medium-sized deletions lead to recognizable human disorders, e.g. Williams syndrome.
Deletion of a number of pairs that is not evenly divisible by three will lead to a frameshift mutation, causing all of the codons occurring after the deletion to be read incorrectly during translation, producing a severely altered and potentially nonfunctional protein. In contrast, a deletion that is evenly divisible by three is called an in-frame deletion.
Deletions are responsible for an array of genetic disorders, including some cases of male infertility, two thirds of cases of Duchenne muscular dystrophy, and two thirds of cases of cystic fibrosis (those caused by ΔF508). Deletion of part of the short arm of chromosome 5 results in Cri du chat syndrome. Deletions in the SMN-encoding gene cause spinal muscular atrophy, the most common genetic cause of infant death.
Microdeletions are associated with many different conditions, including Angelman Syndrome, Prader-Willi Syndrome, and DiGeorge Syndrome. Some syndromes, including Angelman syndrome and Prader-Willi syndrome, are associated with both microdeletions and genomic imprinting, meaning that same microdeletion can cause two different syndromes depending on which parent the deletion came from.
Recent work suggests that some deletions of highly conserved sequences (CONDELs) may be responsible for the evolutionary differences present among closely related species. Such deletions in humans, referred to as hCONDELs, may be responsible for the anatomical and behavioral differences between humans, chimpanzees and other varieties of mammals like ape or monkeys.
Recent comprehensive patient-level classification and quantification of driver events in TCGA cohorts revealed that there are on average 12 driver events per tumor, of which 2.1 are deletions of tumor suppressors.
Detection
The introduction of molecular techniques in conjunction with classical cytogenetic methods has in recent years greatly improved the diagnostic potential for chromosomal abnormalities. In particular, microarray-comparative genomic hybridization (CGH) based on the use of BAC clones promises a sensitive strategy for the detection of DNA copy-number changes on a genome-wide scale. The resolution of detection could be as high as >30,000 "bands" and the size of chromosomal deletion detected could as small as 5–20 kb in length. Other computation methods were selected to discover DNA sequencing deletion errors such as end-sequence profiling.
Mitochondrial DNA deletions
In the yeast Saccharomyces cerevisiae, the nuclear genes Rad51p, Rad52p and Rad59p encode proteins that are necessary for recombinational repair and are employed in the repair of double strand breaks in mitochondrial DNA. Loss of these proteins decreases the rate of spontaneous DNA deletion events in mitochondria. This finding implies that the repair of DNA double-strand breaks by homologous recombination is a step in the formation of mitochondrial DNA deletions.
See also
References

This article uses material from the Wikipedia English article Deletion (genetics), which is released under the Creative Commons Attribution-ShareAlike 3.0 license ("CC BY-SA 3.0"); additional terms may apply (view authors). Content is available under CC BY-SA 4.0 unless otherwise noted. Images, videos and audio are available under their respective licenses.
®Wikipedia is a registered trademark of the Wiki Foundation, Inc. Wiki English (DUHOCTRUNGQUOC.VN) is an independent company and has no affiliation with Wiki Foundation.